Welcome to CRAPome CRAPome
OVERVIEW
CRAPOME.ORG TRAFFIC
Date Range
Date Range
Date Range
CRAPOME.ORG HISTORY
WEBSITE SPAN
LINKS TO WEBSITE
Welcome to the Gingras Lab! We are a signal transduction, systems biology and proteomics lab located in the Lunenfeld-Tanenbaum Research Institute. A Laboratory Information Management System for AP-MS experiments. Explore the Contaminant Repository for Affinity Purification.
Advancing Proteomics, one spectrum at a time. Proteomics and Integrative Bioinformatics Lab. Department of Computational Medicine and Bioinformatics. Director, Proteomics Resource Facility. University of Michigan, Ann Arbor, Michigan.
Francine Bomar, Chief Administrator. Sheryl Hansen Smith, M. Strengthening basic science research and education at the University of Michigan.
ProHits-viz generates a variety of high-quality, customizable images from protein-protein interaction data. Explicit support is provided for output from SAINT. And generic support is provided for output from other tools. The interactive viewers can be used to visualize output from our tools directly in the browser. 169; 2015-2016, the Gingras lab. Link on the connecting page. Link on the connecting page. Link on the connecting page. This tool takes a file with quantitative information on bait-prey intera.
WHAT DOES CRAPOME.ORG LOOK LIKE?

CONTACTS
CRAPOME.ORG SERVER
NAME SERVERS
WEBSITE ICON

SERVER SOFTWARE
We identified that this website is operating the Apache/2.2.15 (Red Hat) server.SITE TITLE
Welcome to CRAPome CRAPomeDESCRIPTION
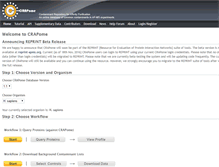
Skip to main content. Announcing REPRINT Beta Release. We are happy to announce that CRAPome will soon be part of the REPRINT Resource for Evaluation of Protein Interaction Networks suite of tools. The beta version is now available at reprint-apms.org. Step 1 Choose Version and Organism. Choose CRAPome Database Version. Your currently selected organism is H. sapiens. Step 2 Choose Workflow. Workflow 1 Query Proteins against CRAPome. Workflow 2 Download Background Contaminant Lists.PARSED CONTENT
The domain crapome.org states the following, "We are happy to announce that CRAPome will soon be part of the REPRINT Resource for Evaluation of Protein Interaction Networks suite of tools." I analyzed that the webpage said " The beta version is now available at reprint-apms." They also stated " Step 1 Choose Version and Organism. Your currently selected organism is H. Workflow 1 Query Proteins against CRAPome. Workflow 2 Download Background Contaminant Lists."ANALYZE OTHER DOMAINS
Added the gay-ass pick of the anime thingie. Well anyway I updated the quote. And lets get the counter to 100 by JUNE! Tell your friends about this site! The new month is about to start. All I had time to do was add the counter and update the quote. I added my first poem.
Subscribe to my blog! Bienvenue sur le Skyblog des. DONNEZ MOI LA PAGE DE VOTRE BLOG AINSI QUE LA PHOTO SOUHAITEE.
Ma Commune a du coeur. La 4G à Craponne sur Arzon. Maison de Service Au Public. Enfance Jeunesse et Centre Social. Agenda des Loisirs et manifestations. Appel à projet de De.